HMG-box
Appearance move to sidebar hide| HMG (high mobility group) box | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
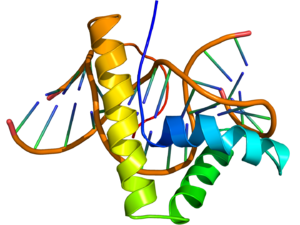 NMR structure of the HMG-box domain of the LEF1 protein (rainbow colored, N-terminus = blue, C-terminus = red) complexed with DNA (brown) based on the PDB: 2LEF coordinates. NMR structure of the HMG-box domain of the LEF1 protein (rainbow colored, N-terminus = blue, C-terminus = red) complexed with DNA (brown) based on the PDB: 2LEF coordinates. | |||||||||||
| Identifiers | |||||||||||
| Symbol | PF00505 | ||||||||||
| Pfam | PF00505 | ||||||||||
| InterPro | IPR009071 | ||||||||||
| SCOP2 | 1hsm / SCOPe / SUPFAM | ||||||||||
| |||||||||||
In molecular biology, the HMG-box (high mobility group box) is a protein domain which is involved in DNA binding. The domain is composed of approximately 75 amino acid residues that collectively mediate the DNA-binding of chromatin-associated high-mobility group proteins. HMG-boxes are present in many transcription factors and chromatin-remodeling complexes, where they can mediate non-sequence or sequence-specific DNA binding.
Structure
The structure of the HMG-box domain contains three alpha helices separated by loops (see figure to the right).
Function
HMG-box containing proteins only bind non-B-type DNA conformations (kinked or unwound) with high affinity. HMG-box domains are found in some high mobility group proteins, which are involved in the regulation of DNA-dependent processes such as transcription, replication, and DNA repair, all of which require changing the conformation of chromatin. The single and the double box HMG proteins alter DNA architecture by inducing bends upon binding.
References
- ^ a b Stros M, Launholt D, Grasser KD (October 2007). "The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins". Cell. Mol. Life Sci. 64 (19–20): 2590–606. doi:10.1007/s00018-007-7162-3. PMC 11136187. PMID 17599239. S2CID 28156847.
- ^ Štros, M.; Launholt, D.; Grasser, K. D. (October 2007). "The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins". Cellular and Molecular Life Sciences. 64 (19–20): 2590–2606. doi:10.1007/s00018-007-7162-3. PMC 11136187. PMID 17599239. Retrieved 18 February 2023.
- ^ a b Thomas JO (August 2001). "HMG1 and 2: architectural DNA-binding proteins". Biochem. Soc. Trans. 29 (Pt 4): 395–401. doi:10.1042/BST0290395. PMID 11497996.
- ^ D. Murugesapillai et al, DNA bridging and looping by HMO1 provides a mechanism for stabilizing nucleosome-free chromatin, Nucleic Acids Res (2014) 42 (14): 8996-9004
- ^ D. Murugesapillai et al, Single-molecule studies of high-mobility group B architectural DNA bending proteins, Biophys Rev (2016) doi:10.1007/s12551-016-0236-4
External links
- HMG-Box Domains at the U.S. National Library of Medicine Medical Subject Headings (MeSH)
This protein-related article is a stub. You can help Wikipedia by expanding it. |